On January 27, 2024, our research group and the reserach group of Professor Yiyue Zhang from South China University of Technology jointly published a research paper titled "Cebp1 and Cebpβ transcriptional axis controls eosinophilopoiesis in zebrafish" in Nature Communications, revealing the spatiotemporal development patterns, lineage characteristics and regulatory mechanisms of eosinophils using the zebrafish model.

Previous understanding of eosinophil development and regulation mainly comes from adult hematopoiesis in mammalian models: functional eosinophils are determined by bone marrow-derived eosinophils to differentiate and mature progenitor cells (EoP) step by step [1]. In this study, we used a zebrafish model with a hematopoietic system very similar to that of mammals to analyze the developmental characteristics and regulatory mechanisms of eosinophils from early development to adulthood. First, we identified the specific molecular marker eslec of zebrafish eosinophil lineage, constructed the transgenic reporter line Tg (eslec: eGFP) zebrafish lineage, and performed overall imaging of eosinophil development, and found that eosinophils began to be produced and gradually increased 5 days after birth in zebrafish, and were mainly distributed in renal marrow (equivalent to mammalian bone marrow), peritoneal fluid, intestine and other tissues, showing the early spatiotemporal development pattern of eosinophils. Subsequently, we used single-cell sequencing technology (ScRNA-Seq) to divide the eosinophil lineage into progenitor cells (EoP), precursor cells (pre. Eos) and mature cells (mat.Eos), and analyzed the lineage characteristics of eosinophils: EoP had relatively active proliferation. pre.Eos mainly enriched ribosomes, protein modifications and oxidative phosphorylation pathways, which may be related to a large number of synthetic granulin proteins. mat. EOS expresses more immune-related genes. Then, we further explored the hierarchical regulatory mechanism of Cebp molecule on eosinophil lineage development, and compared the conservation and differences between Cebp regulatory granule hematopoiesis species. The results revealed the fine mechanism of the Cebp1-Cebpβ transcriptional regulatory axis in the development of eosinophil lineage: Cebp1 can inhibit the lineage determination of eosinophils and may affect the differentiation of precursor cells. Cebpβ promotes lineage determination of eosinophils and pre.eos differentiation. The study also found that the human C/EBPeP27 protein has a similar function to zebrafish Cebp1 and can inhibit the development of eosinophils in zebrafish, and zebrafish Cebp1 and human C/EBPeP27 are conserved in regulating eosinophil development.
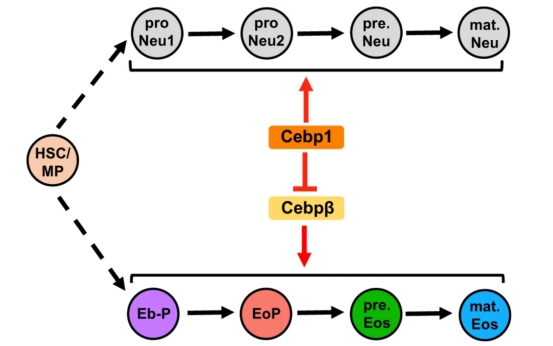
Schematic diagram of the content of the study
In conclusion, this study identified the specific molecular marker ESLEC of zebrafish eosinophils, established a lineage-specific transgenic reporter line, analyzed the spatiotemporal development pattern of eosinophils and their lineage stage developmental characteristics, discovered the regulatory mechanism of the Cebp1-Cebpβ regulatory axis in eosinophil lineage development, and revealed the conservation and difference between Cebp1 and C/EBPe on the regulation of vertebrate granulosion hematopoiesis. The study revealed the spatiotemporal development patterns, lineage characteristics and regulatory mechanisms of eosinophils. Dr. Gaofei Li and PhD student Yicong Sun from the School of Medicine of South China University of Technology are the co-first authors of the paper, and Professor Zhang Yiyue and Professor Lai Guan Ng from South China University of Technology are the co-corresponding authors of the paper. Professor Florent Ginhoux and Dr. Immanuel Kwok from the Singapore Collaborative Institute on Immunology, Researcher Liu Zhaoyuan from the Shanghai Institute of Immunology, and Dr. Pei Jinxin, Dr. He Shuai and Dr. Peng Wan from the Cancer Prevention and Treatment Center of Sun Yat-sen University also participated in this study. This paper is supported by the key research and development project of the National Key R&D Program "Developmental Programming and Metabolic Regulation" and the National Natural Science Foundation of China. Link to the paper:https://www.nature.com/articles/s41467-024-45029-0 References:
1. P.C. Fulkerson, Transcription Factors in Eosinophil Development and As Therapeutic Targets, Front. Med. 4 (2017) 1–6. doi:10.3389/fmed.2017.00115.
|